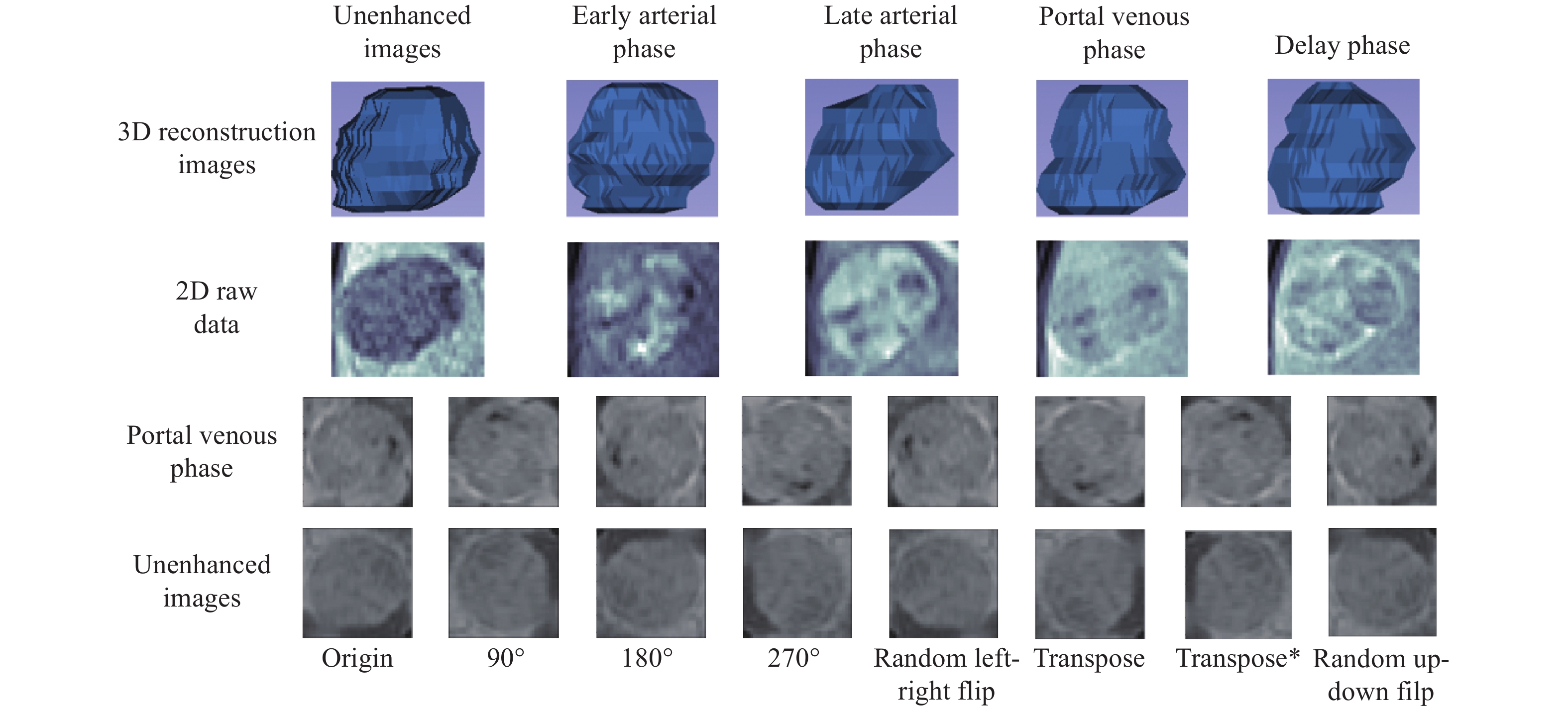
Self-attention guided multi-sequence fusion model for differentiation of hepatocellular carcinoma
-
摘要: 結合影像學和人工智能技術對病灶進行無創性定量分析是目前智慧醫療的一個重要研究方向。針對肝細胞癌(Hepatocellular carcinoma, HCC)分化程度的無創性定量估測方法研究,結合放射科醫師的臨床讀片經驗,提出了一種基于自注意力指導的多序列融合肝細胞癌組織學分化程度無創判別計算模型。以動態對比增強核磁共振成像(Dynamic contrast-enhanced magnetic resonance imaging, DCE-MRI)的多個序列為輸入,學習各時序序列及各序列的多層掃描切片在分化程度判別任務的權重,加權序列中具有的良好判別性能的時間和空間特征,以提升分化程度判別性能。模型的訓練和測試在三甲醫院的臨床數據集上進行,實驗結果表明,本文所提出的肝細胞癌分化程度判別模型取得相比幾種基準和主流模型最高的分類計算性能,在WHO組織學分級任務中,判別準確度、靈敏度、精確度分別達到80%,82%和82%。
-
關鍵詞:
- 肝細胞癌分級 /
- 動態對比增強核磁共振成像 /
- 自注意力機制 /
- 多序列融合 /
- 輔助診斷
Abstract: Hepatocellular carcinoma (HCC) is a type of primary malignant tumor and an urgent problem to be solved, particularly in China, one of the countries with the highest prevalence of HCC. In the choice of treatment methods for patients with hepatocellular carcinoma, accurate histological grading of the lesion area undoubtedly plays a vital role that helps the management and therapy of HCC patients. However, the current pathological detection as the gold standard has defects, such as invasiveness and a large sampling error. Therefore, it is an important direction of intelligent medical treatment to provide noninvasive and accurate lesion grading using imaging technology combined with artificial intelligence technology. On the basis of the radiologists' experience in reading clinical images, this paper proposed a self-attentional guidance-based histological differentiation discrimination model combined with multi-modality fusion and an attention weight calculation scheme for dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) sequences of hepatocellular carcinoma. The model combined the spatiotemporal information contained in the enhancement sequence and learned the importance of each sequence and the slice in the sequence for the classification task. It effectively used the feature information contained in the enhancement sequence in the temporal and spatial dimensions to improve the classification performance. During the experiment, the model was trained and tested on the clinical data set of the top three hospitals in China. The experimental results show that the self-attention-guided model proposed in this paper achieves higher classification performance than several benchmark and mainstream models. Comprehensive experiments were performed on the clinical dataset with labels annotated by professional radiologists. The results show that our proposed self-attention model can achieve acceptable quantitative measuring of HCC histologic grading based on the MRI sequences. In the WHO histological classification task, the classification accuracy of the proposed model reaches 80%, the sensitivity is 82%, and the precision is 82%. -
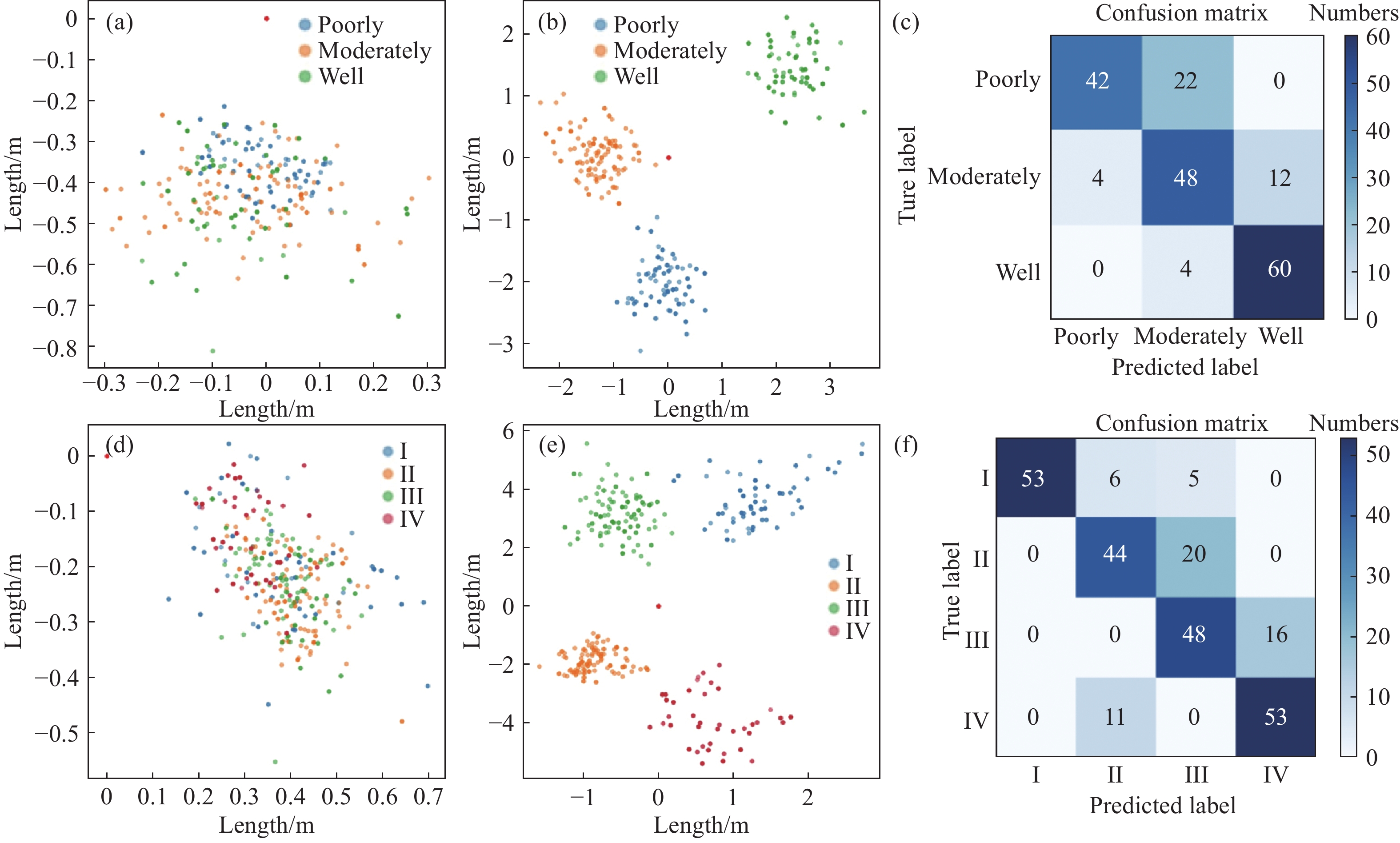
圖 3 HCC三分類和四分類任務中“自注意力”模型的embedding space和混淆矩陣。(a)三分類任務訓練前的特征空間;(b)三分類任務訓練后的特征空間;(c)三分類任務的混淆矩陣;(d)四分類任務訓練前的特征空間;(e)四分類任務訓練后的特征空間;(f)四分類任務的混淆矩陣
Figure 3. Feature distributions at the embedding space before and after training and the corresponding confusion matrix of the WHO and Edmonson classification tasks: (a) feature space of the model in three classification tasks before training; (b) feature space of the model in three classification tasks after training; (c) confusion matrix in three classification tasks; (d) feature space of the model in four classification tasks before training; (e) feature space of the model in four classification tasks after training; and (f) confusion matrix in four classification tasks
表 1 基于WHO分類標準的HCC類別數據分布
Table 1. Augmentation results for a dataset with HCC grading under the WHO grading system
Datasets Well Moderately Poorly Training set 56 208 54 Test set 32 104 26 Total 88 312 80 表 2 基于Edmonson分類標準的HCC類別數據分布
Table 2. Augmentation results for a dataset with HCC grading under the Edmonson grading system
Datasets I II III IV Training set 56 88 120 54 Test set 32 40 64 26 Total 88 128 184 80 表 3 基于WHO分類標準的對比實驗
Table 3. Detailed comparison of experimental results on the test set under the WHO grading standard
Model Accuracy Recall Precision F1-score Our method 0.8021±0.0478 0.8231±0.0404 0.8215±0.0537 0.8221±0.0477 MCF-3DCNN[15] 0.7188±0.0405 0.6667±0.0195 0.7874±0.0416 0.7014±0.0337 3D ResNet[22] 0.7312±0.0627 0.7353±0.0557 0.7762±0.0443 0.7613±0.0537 3D SE-ResNet[23] 0.7453±0.0675 0.7342±0.0755 0.7627±0.0732 0.7665±0.0675 3D SE-DenseNet[24] 0.7854±0.0445 0.7923±0.0638 0.8117±0.0417 0.7913±0.0576 中文字幕在线观看表 4 基于Edmonson分類標準的對比實驗
Table 4. Detailed comparison of experimental results on the test set under the Edmonson grading standard
Model Accuracy Recall Precision F1-score Our method 0.7734±0.0318 0.7889±0.0412 0.8089±0.0416 0.7896±0.0225 MCF-3DCNN[15] 0.6322±0.0522 0.5482±0.1338 0.6424±0.0657 0.6431±0.0824 3D ResNet[22] 0.7037±0.0731 0.7229±0.0442 0.7404±0.0421 0.7203±0.0336 3D SE-ResNet[23] 0.7108±0.0644 0.7492±0.0531 0.7637±0.0788 0.7566±0.0631 3D SE-DenseNet[24] 0.7227±0.0341 0.7762±0.0426 0.7738±0.0446 0.7876±0.0512 -
參考文獻
[1] Yang J D, Hainaut P, Gores G J, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol, 2019, 16(10): 589 doi: 10.1038/s41575-019-0186-y [2] Jiang Y, Sun A H, Zhao Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature, 2019, 567(7747): 257 doi: 10.1038/s41586-019-0987-8 [3] Lin H X, Wei C, Wang G X, et al. Automated classification of hepatocellular carcinoma differentiation using multiphoton microscopy and deep learning. J Biophoto, 2019, 12(7): e201800435 [4] Jimenez H, Wang M H, Zimmerman J W, et al. Tumour-specific amplitude-modulated radiofrequency electromagnetic fields induce differentiation of hepatocellular carcinoma via targeting Cav3.2 T-type voltage-gated calcium channels and Ca2+ influx. EBioMedicine, 2019, 44: 209 doi: 10.1016/j.ebiom.2019.05.034 [5] Shioga T, Kondo R, Ogasawara S, et al. Usefulness of tumor tissue biopsy for predicting the biological behavior of hepatocellular carcinoma. Anticancer Res, 2020, 40(7): 4105 doi: 10.21873/anticanres.14409 [6] Parr R L, Mills J, Harbottle A, et al. Mitochondria, prostate cancer, and biopsy sampling error. Discov Med, 2013, 25;15(83): 213 [7] Henken K, Van Gerwen D, Dankelman J, et al. Accuracy of needle position measurements using fiber Bragg gratings. Minim Invasive Ther Allied Technol, 2012, 21(6): 408 doi: 10.3109/13645706.2012.666251 [8] Li J Z, Xue F, Xu X H, et al. Dynamic contrast enhanced MRI differentiates hepatocellular carcinoma from hepatic metastasis of rectal cancer by extracting pharmacokinetic parameters and radiomic features. Exp Ther Med, 2020, 20(4): 3643 [9] Kaissis G A, Loh?fer F K, H?rl M, et al. Combined DCE-MRI- and FDG-PET enable histopathological grading prediction in a rat model of hepatocellular carcinoma. Eur J Radiol, 2020, 124: 108848 doi: 10.1016/j.ejrad.2020.108848 [10] Khalifa F, Soliman A, El-Baz A, et al. Models and methods for analyzing DCE-MRI: A review. Med Phys, 2014, 41(12): 124301 doi: 10.1118/1.4898202 [11] Yang D W, Jia X B, Xiao Y J, et al. Noninvasive evaluation of the pathologic grade of hepatocellular carcinoma using MCF-3DCNN: A pilot study. Biomed Res Int, 2019: 9783106 [12] Chernyak V, Fowler K J, Kamaya A, et al. Liver imaging reporting and data system (LI-RADS) version 2018: Imaging of hepatocellular carcinoma in at-risk patients. Radiology, 2018, 289(3): 816 doi: 10.1148/radiol.2018181494 [13] Suk H I, Lee S W, Shen D G. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. NeuroImage, 2014, 101: 569 doi: 10.1016/j.neuroimage.2014.06.077 [14] Wang Q Y, Que D S. Staging of hepatocellular carcinoma using deep feature in contrast-enhanced MR images//2nd International Conference on Computer Engineering, Information Science & Application Technology (ICCIA 2017). Wuhan, 2016: 186 [15] Jia X B, Xiao Y J, Yang D W, et al. Temporal-spatial feature learning of dynamic contrast enhanced-MR images via 3D convolutional neural networks//Chinese Conference on Image and Graphics Technologies. Singapore, 2018: 380 [16] Jia X B, Xiao Y J, Yang D W, et al. Multi-parametric MRIs based assessment of Hepatocellular Carcinoma Differentiation with Multi-scale ResNet. TIIS, 2019, 13(10): 5179 [17] Antropova N, Huynh B Q, Giger M L. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys, 2017, 44(10): 5162 doi: 10.1002/mp.12453 [18] Hu Q Y, Whitney H M, Giger M L. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci Rep, 2020, 10: 10536 doi: 10.1038/s41598-020-67441-4 [19] Zhang T H, Fan S L, Guo X X, et al. Intelligent medical assistant diagnosis method based on data fusion. Chin J Eng, doi: 10.13374/j.issn2095-9389.2021.01.12.003張桃紅, 范素麗, 郭徐徐, 等. 基于數據融合的智能醫療輔助診斷方法. 工程科學學報, doi: 10.13374/j.issn2095-9389.2021.01.12.003 [20] Ye H, Chen Q J, Wu H M, et al. Classification of liver cancer images based on deep learning//International conference on Data Science, Medicine and Bioinformatics. Singapore, 2020: 184 [21] Zhou L, Rui J G, Zhou W X, et al. Edmondson-Steiner grade: A crucial predictor of recurrence and survival in hepatocellular carcinoma without microvascular invasio. Pathol Res Pract, 2017, 213(7): 824 doi: 10.1016/j.prp.2017.03.002 [22] He K M, Zhang X Y, Ren S Q, et al. Deep residual learning for image recognition//2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, 2016: 770 [23] Hu J, Shen L, Sun G. Squeeze-and-excitation networks//2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Salt Lake City, 2018: 7132 [24] Zhou Q, Zhou Z Y, Chen C M, et al. Grading of hepatocellular carcinoma using 3D SE-DenseNet in dynamic enhanced MR images. Comput Biol Med, 2019, 107: 47 doi: 10.1016/j.compbiomed.2019.01.026 [25] Yoshinobu Y, Iwamoto Y, Xianhua H A N, et al. Deep learning method for content-based retrieval of focal liver lesions using multiphase contrast-enhanced computer tomography images//2020 IEEE International Conference on Consumer Electronics (ICCE). Las Vegas, 2020: 1 -



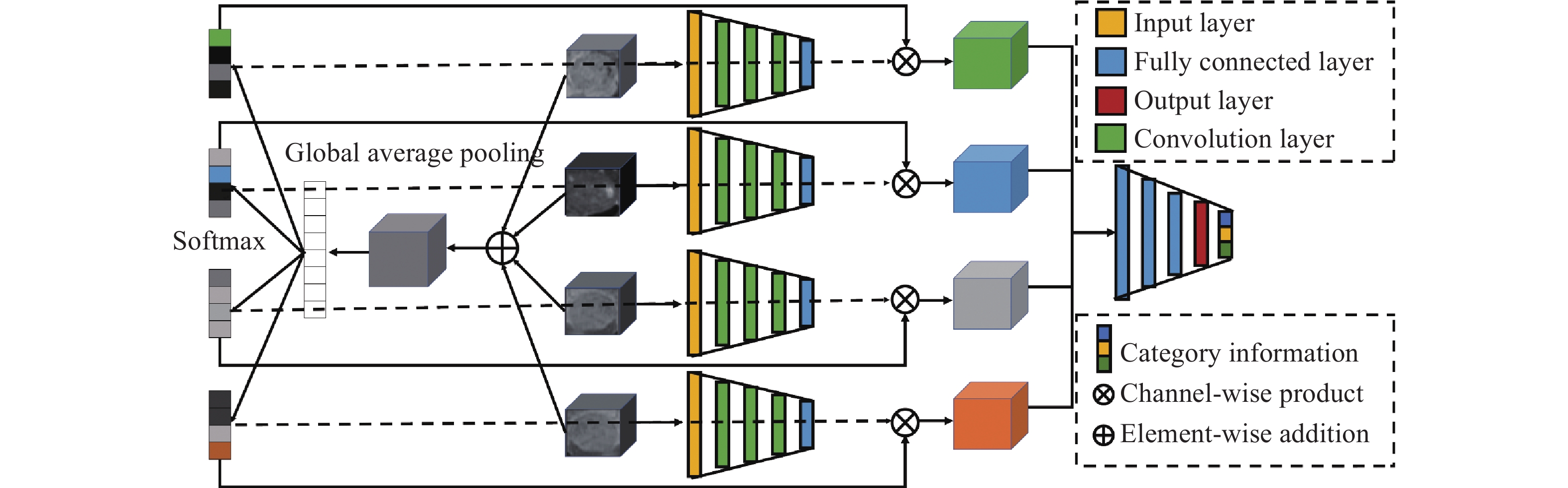
 下載:
下載: